The Community Edition of GeneTegra RDW enables the integration of clinical or phenotype data with sequence data from microorganisms collected from human or animal specimens, such as bacteria, viruses, fungi, archaea and protists. This edition provides the RDW with a dimensional warehouse model for sequence data from microorganisms and host phenotypes, an editable system ontology, and a set of configurable data extraction queries specifically designed for the needs of the infectious diseases epidemiology community. The dimensional warehouse model has been designed under a research project with the Division of Viral Hepatitis at the Centers of Disease Control and Prevention (CDC), and with the participation and active collaboration of expert researchers in viral and microbial epidemiology.
Warehouse Model
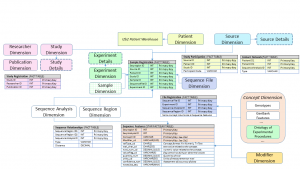
A sequence data warehouse model has been designed to integrate microorganism sequence data and its associated metadata and clinical data, and store it in a central repository suitable for data analysis. The data model uses an entity-attribute-value (EAV) star schema appropriate for sequence data with sparse meta-data annotations and patient data with sparse attributes. It involves two main transactional data sources, a patient and a sequence data source, used to store the data generated, downloaded or collected on an ongoing basis. It also includes an external GeneTegra RDW ontology containing the reference information that gives context to the transactional data. This allows the user to retrieve the data based on a subset of attributes, and to run data mining studies on it.
System Ontology
The GeneTegra RDW system ontology for infectious diseases includes concepts from the domain of medical microbiology, infectious disease outbreak, transmission, and surveillance studies. As an example, the input data that needs to be stored in the warehouse may contain information about the results of an HCV serology lab test. If the different types of tests and the possible results have not been configured, these concepts can be added to the ontology through the Ontology Management web interface.
The ontology includes concepts specific to patients and the microorganisms sampled from those patients and concepts specific to DNA/RNA sequence data. The patients ontology includes the usual demographic and clinical concepts (such as ICD-10), as well as concepts specific to epidemiology and treatment data. For sequence annotations, concept codes were considered in agreement with the new GSCID/BRC sequence metadata standards published by the NIAID in 2014, which covers the information required to submit data to NCBI BioProject and BioSample. Concepts that apply to Next Generation Sequencing (NGS) and analysis, experimental details, and file and sequence read statistics, are also included.
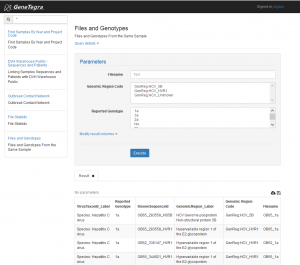
Data Search and Export
GeneTegra RDW uses the GeneTegra Web Query interface to search for data. A list of queries is made available that link different datasets together such as patient demographics to patient observations, treatments and medications, and to blood specimens and DNA or RNA sequences. Query search results can be exported to delimiter separated files.